Compare
We compare the innovative Dot-it-Spot-it Total Protein Assay with two established assays, examining sensitivity, specificity, compatibility, and protein-to-protein variation.
Sensitivity
Specificity in biological specimens
Specificity and compatibility
Protein-to-protein variation
SENSITIVITY
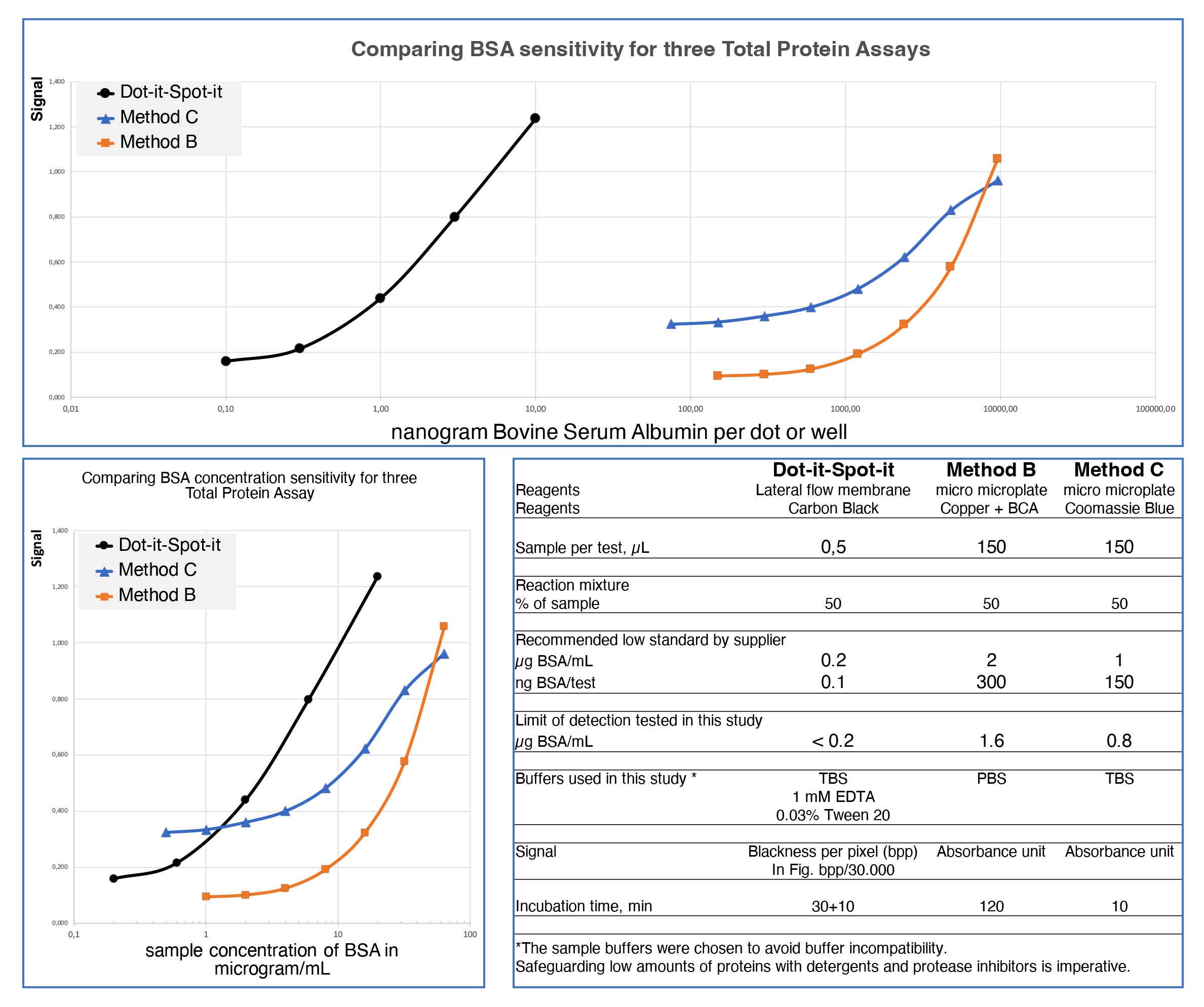
Sensitivity and sample volume
This study compares the most sensitive variants of three Total Protein Assays to determine their suitability for quantifying protein in minute details of valuable biological samples.
The Dot-it-Spot-it Total Protein Assay was most suitable for this purpose, exhibiting 2000 times greater sensitivity and measuring low protein concentrations using only 0.5 microlitres of the sample.
SPECIFICITY in biological specimens
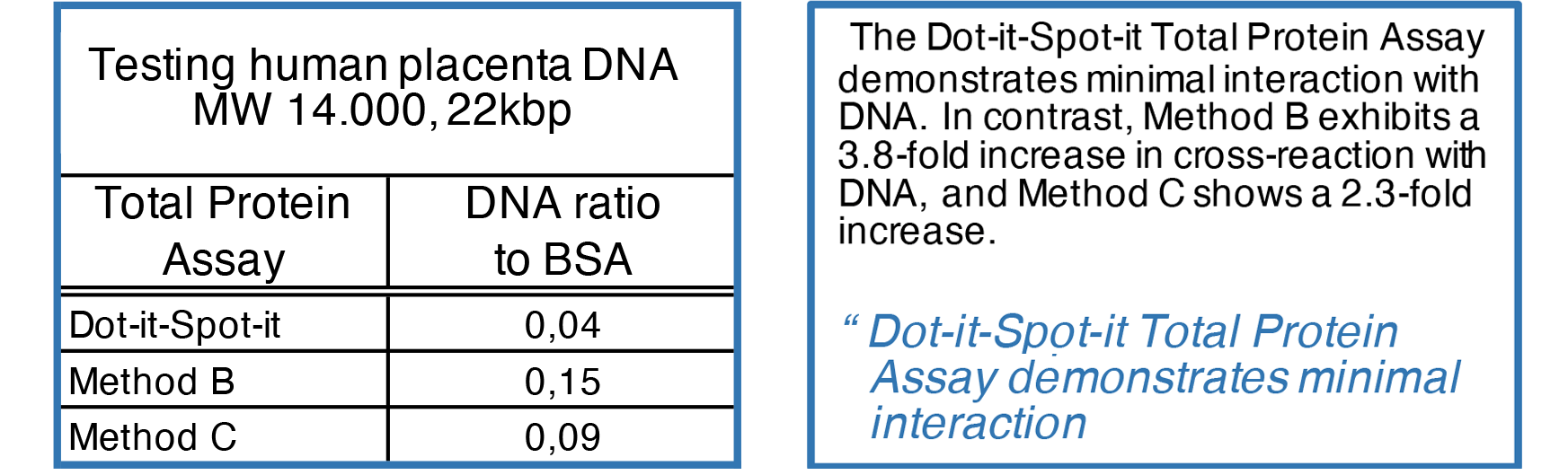
DNA Cross-Reaction
While total protein assays are effective in measuring protein content, they may also exhibit cross reactions with other biological molecules, such as DNA. These assays can display varying degrees of crossreaction. For instance, in a sample containing 100 μg of DNA, a false-positive result ranging from 15 to 4 μg of BSA could be observed.
SPECIFICITY and COMPATIBILITY
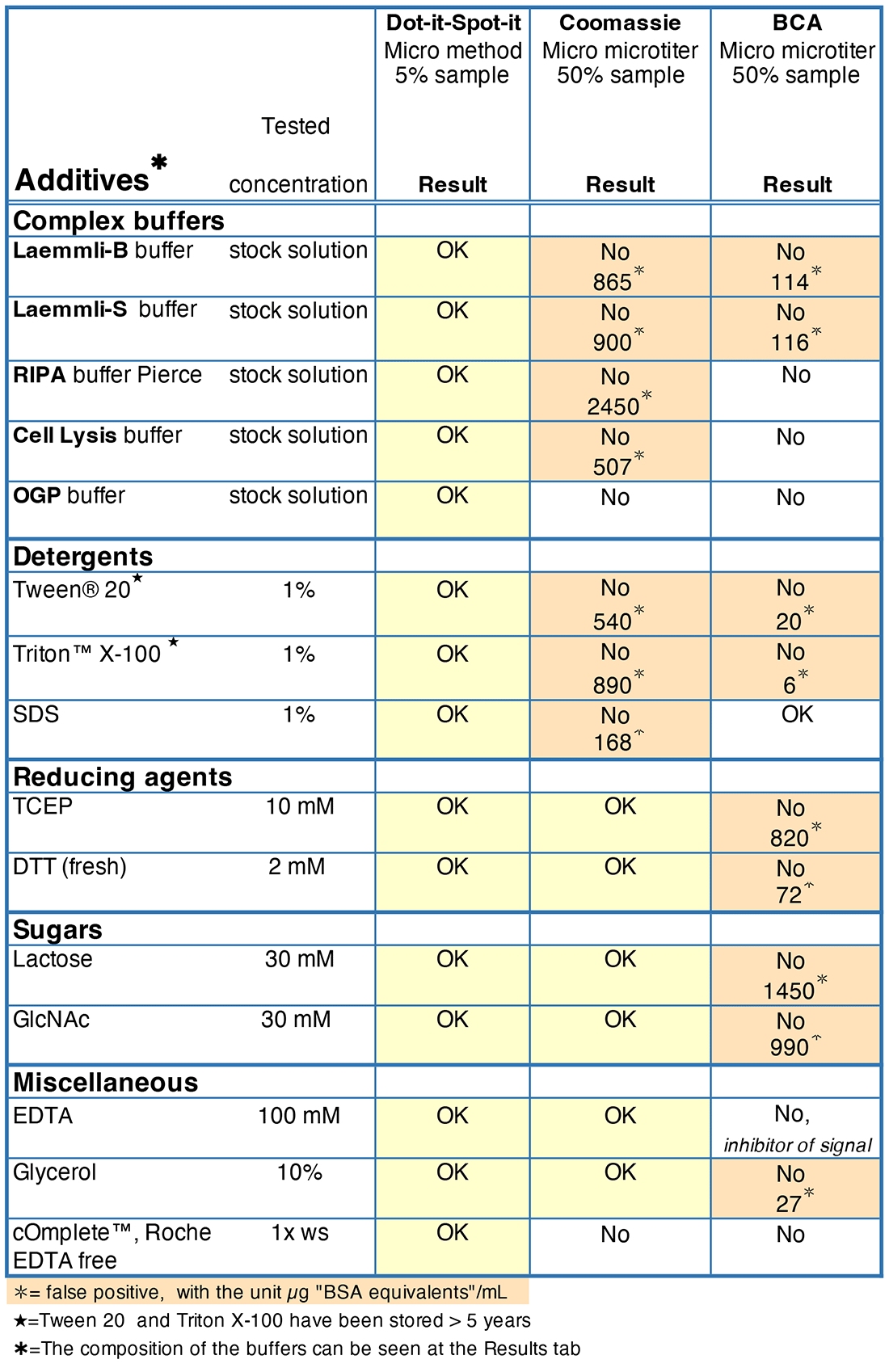
Incompatibility with essential additives when working with low amounts of proteins:
When protein concentration is low, more sample is required for analysis, resulting in larger issues with incompatibility of buffer additives.
The table below shows three methods that can measure down to 2±1 μg BSA/mL in TBS or PBS buffer. However, successful handling of proteins requires additives shown in the table to extract, dissolve, and store them.
The ultra-sensitive Dot-it-Spot-it Total Protein Assay experiences significantly less interference from additives and provides greater sensitivity to proteins compared to current assays.
PROTEIN-TO-PROTEIN VARIATION
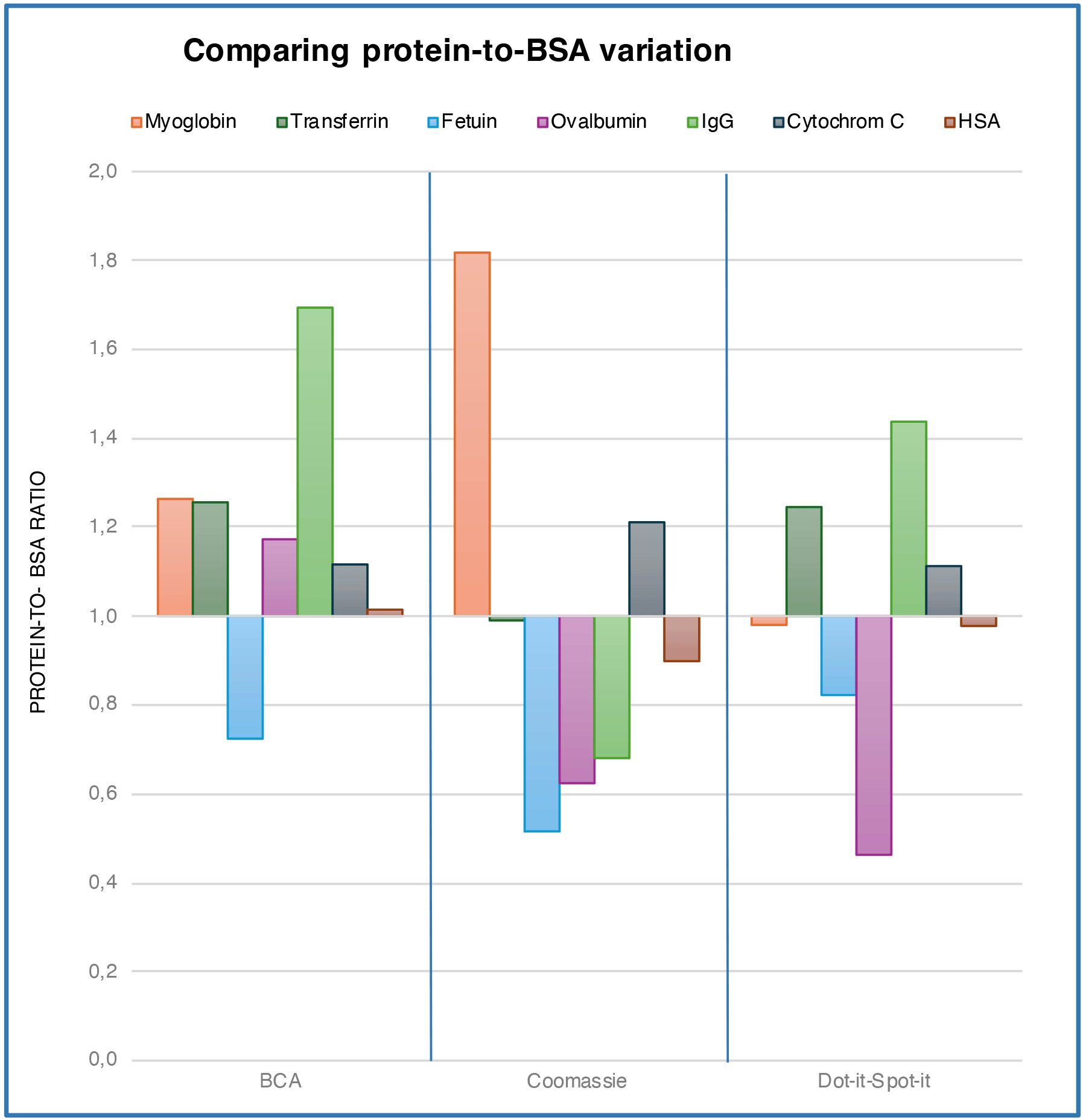
The protein-to-BSA ratio
Proteins vary in their amino acid composition and sequence, which directly influences their structure. Moreover, proteins can undergo various PTMs, such as phosphorylation, glycosylation, acetylation, and ubiquitination, further diversifying their structure.
These variations affect Total Protein Assays differently. The novel Dot-it Spot-it method interacts with proteins similarly to the two conventional tests.
For further information, please refer to the ‘Results’ tab.
The novel Dot-it-Spot-it method interacts with proteins similarly to the two conventional tests